
Optimization of laying hens breeding programs
WIAS Magazine - Winter edition 2023
Research Overview
In 2022, with Germany and Spain, France was among the leading egg producers in Europe. Around 14 billion eggs were produced, representing 14% of the European production (1). This production has been steadily increasing for years, meeting the growing demand from consumers. The egg, being a cost-effective source of animal protein compared to other animal products (€4.2/100g of proteins), plays a significant role in this trend. Behind dairy farming, egg-laying hen farming is the most environmentally friendly in terms of greenhouse gas emissions, emitting less than 2kg of CO2 per kilogram of eggs produced. Moreover, eggs are a universally accepted food item across different dietary preferences and are easy to store. In 2022, an average French person consumed 4.3 eggs per week resulting in a 0.3% increase compared to the previous year (2). Selective breeding plays a crucial role in supporting the increasing production needs of the industry.
The egg sector is organized in a pyramid structure with the breeding companies at the top that select animals from purebred lines (Figure 1). Some individuals from purebred lines (at the top of the pyramid) are mated across lines over several generations to produce crossbred individuals, which are then placed in commercial farms for egg production. The other part of the purebred line individuals constitutes the breeding stock. After their rearing period, usually in individual cages, these selection candidates are phenotyped and their breeding values are estimated using pedigree, phenotypic information and eventually genotypes. The best candidates (for the EBVs) are selected and mated to produce the next generation of the breeding stock. The aim of the selection scheme is to increase genetic gain, generation after generation, and to disseminate this progress to the lower stages of the pyramid.
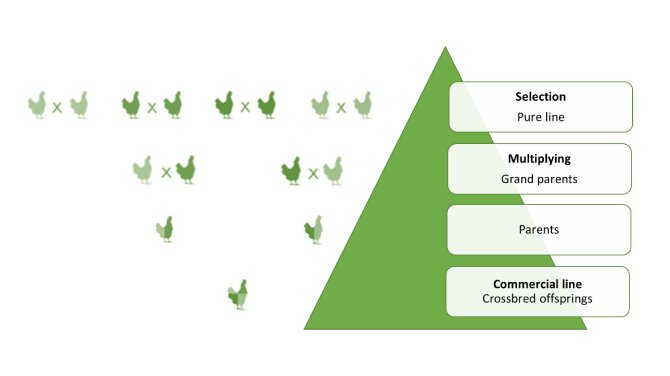
Figure 1: Pyramidal structural organization of poultry breeding companies
The global aim of my PhD is to optimize the features of breeding programs so they can fit tomorrow’s challenges. The first part of my PhD is dedicated to the optimization of the genetic evaluation phase. Indeed, since the 2010s, genomic information has been routinely used to evaluate selection candidates. By using their genotypes, the genetic potential of the candidates can be evaluated. Thanks to a training population that is genotyped and phenotyped, it is possible to estimate the effect of each genotype of a DNA maker on the phenotype. Then, for a related testing population, only genotypes are needed to estimate breeding values. Genotyping the selection candidates ensures a better accuracy of the relationships between individuals and therefore, a more accurate estimation of their breeding values. Genomic selection also allows for the selection of individuals earlier in their career. This results in a more efficient increase in genetic gain over time with genomic selection than with pedigree-based selection (3). However, high-density (HD) genotyping of all selection candidates can be costly for breeding companies, particularly in the poultry industry where the economic value of breeding animals is very low. One of the major challenges for breeding companies today is to reduce these genotyping costs without compromising the accuracy of evaluations. Then, usually, breeding companies select only a portion of the markers of the HD chip and create low-density (LD) chips adapted to their lines, which are cheaper. This solution has some drawbacks. First, not all the chromosomes known are covered by the original HD chip, meaning that these are also missing from the LD chip. In addition, chip prices are highly dependent on the companies selling them and the volume ordered.
Consequently, we decided to test an alternative method for the genotyping and the genomic evaluation of layers: the ddRAD-seq. It is a reduced-representation genome sequencing approach that appeared, to be a plausible alternative to the HD or LD chips, based on simulation studies conducted by Herry et al. in 2019 (4). The method relies on a digestion of DNA with a combination of two restriction enzymes. Only enzymatic fragments generated by the combination of the two restriction enzymes are sequenced (5). This way, the genotyping markers common to all individuals in the population are well distributed across the genome. The potential of this method for the genomic selection of our laying hen line must therefore be estimated.
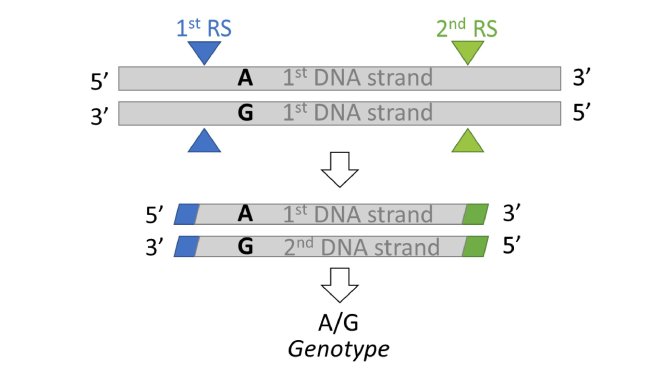
Figure 2: ddRAD-seq principle. DNA is digested with 2 different restriction enzymes. They cut DNA at specific loci on the genome called restriction sites (RS). Then, only enzymatic fragments generated by the combination of the two restriction enzymes are sequenced. SNP located on these enzymatic fragments are genotyped.
In addition to the switch to genomic selection, the laying hen industry is facing structural changes: since 2017, a European movement called End the cage age has initiated an end to the rearing of animals in cages. In France, the consequences of this movement have quickly become apparent. In 2022, 77% of French laying hens were reared on the ground (CNPO), and this proportion continues to rise. There is also a desire to lengthen laying hen’s careers as much as possible to raise fewer animals and simultaneously ensure production. Therefore, breeding companies have to adapt their breeding objectives, breeding program and the breeding farms themselves, to accommodate these changes.
Therefore, the second part of my PhD will deal with the optimization of genomic selection programs in relation to alternative breeding systems. As all animals will have to be bred and mated on the ground, we need to design systems where males and females are bred in the same areas. We will have less control over mating and as a result on the pedigree. We will also have to consider a higher mortality rate in alternative breeding than in cages. New egg-laying persistence traits will have to be added to the selection objectives. I will investigate different breeding programs in the form of simulations to find an optimal breeding strategy given the new circumstances. All the simulations will be carried out with the R package MoBPS (6). Breeding schemes will be assessed using a synthetic indicator taking into account the development of the genetic gain in relation to the development of the population inbreeding rate. The objective is to maximize the genetic gain while minimizing the inbreeding in the population.
Finally, the last part of my PhD will focus on the economic optimization of the most promising breeding programs, using alternative genotyping approaches to chips. The last objective of PhD is to build efficient breeding programs for the selection of laying hens.
In conclusion, the first objective of my PhD is to provide scientific knowledge on the impact of changing breeding and selection systems for laying hens. The second objective is to propose ways of optimizing breeding methods technically and economically, so that they can adapt to these changes in systems to come. For me, this PhD is a great opportunity to gain expertise in a variety of fields crucial to the future of the livestock industry, which I want to participate in.
1. CNPO Site filière [Internet]. [cité 5 avr 2022]. Les chiffres clés. Disponible sur: https://oeuf-info.fr/infos-filiere/les-chiffres-cles/
2. ITAVI, Filière poule pondeuse [Internet]. 2022 [cité 26 févr 2024]. Disponible sur: https://www.itavi.asso.fr/page/filiere-poules-pondeuses
3. Meuwissen THE, Hayes BJ, Goddard ME. Prediction of Total Genetic Value Using Genome-Wide Dense Marker Maps. Genetics. 1 avr 2001;157(4):1819‑29.
4. Herry F, Hérault F, Lecerf F, Lagoutte L, Doublet M, Picard-Druet D, et al. Restriction site-associated DNA sequencing technologies as an alternative to low-density SNP chips for genomic selection: a simulation study in layer chickens. BMC Genomics. 2023;24(1):271.
5. Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double Digest RADseq: An Inexpensive Method for De Novo SNP Discovery and Genotyping in Model and Non-Model Species. PLOS ONE. 31 mai 2012;7(5):e37135.
6. Pook T, Schlather M, Simianer H. MoBPS - Modular Breeding Program Simulator. G3 GenesGenomesGenetics. 1 juin 2020;10(6):1915‑8.




