Project
Knowledge-based prospects for recovery of the Caribbean long-spined sea urchin Diadema antillarum: Understanding mechanisms behind the short- & long-term effects of the 2022 die-off event.
Motivation:
The long-spined sea urchin, Diadema antillarum is a key herbivore on Caribbean reefs. It feeds on a variety of macroalgae, thereby creating free space and facilitating coral recruitment. In 1983 a large die-off event occurred, reducing D. antillarum population densities by on average of 98%. The sudden decrease in grazing pressure, followed by a very limited D. antillarum recovery, resulted in a persistent phase shift from coral-dominated towards algae-dominated reefs in the Caribbean. Currently, multiple organizations are trying to restore D. antillarum populations in order to rehabilitate coral reefs from excessive macroalgal overgrowth. However, in January 2022 a new die-off event occurred, quickly spreading across the Caribbean and again leading to high mortality rates. The causative agent for the 2022 die-off event was identified to be a scuticociliate (associated with Philasterapodigitiformis), but how it causes mortality remains unknown as well as why other sea urchin species in the same ecosystems were not affected. Our research aims to elucidate the mechanism behind scuticociliate infection and disease process. This knowledge is crucial to predict the occurrence and trajectory of future die-off events, and the potential for D. antillarum population establishment by natural recovery or active restocking.
Aim & objectives/research questions:
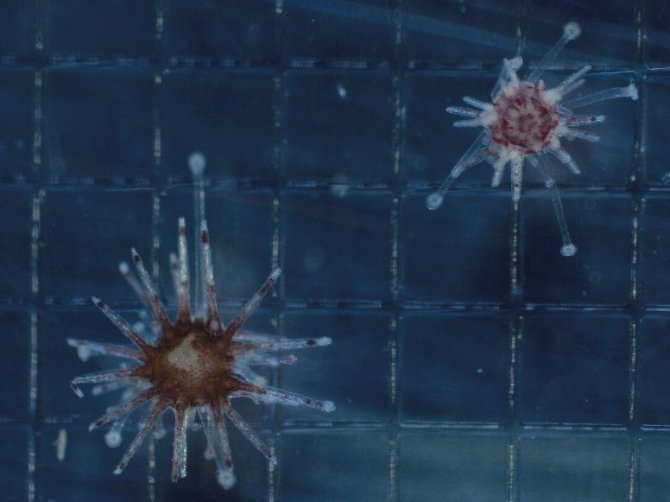
Work package 1: Understanding how Philaster sp. affects mechanisms associated with survival in D. antillarum
- RQ1) What is the effect of sp. on the health and survival of different life stages of ?
- RQ2) How are mechanisms associated with survival of sp. exposure different between unaffected (Caribbean) sea urchin species compared to ?
- RQ3) What is the genetic diversity of populations in the Caribbean?
- RQ4) What are the sink- and source populations of in the Caribbean?
- RQ5) How is population recovery of affected by pathogen outbreaks and changes in population connectivity?
Methodology:
Work package 1: Survival mechanisms towards Philaster sp.
Philaster sp. was identified as the causative agent of the 2022 die-off event, but how Philaster sp. causes disease is not yet known. Additionally, only adult D. antillarum were studied. Currently it is unknown at which life stages Philaster sp. can cause infection in D. antillarum. For example, D. antillarum has a pelagic larval phase in which they stay in the water column for at least 30 days. During that time, the larvae are able to be dispersed through water currents over large distances, making this life stage crucial for population connectivity and gene flow. If the larvae could carry, or would be affected by Philaster sp., this would pose an extra risk for the population. Therefore this PhD project will study the effect of Philaster sp. on the health and survival of different life stages of D. antillarum in an effort to better understand how infection occurs and how it induced disease. I will also investigate cellular immune responses and gene expression before and during infection to see how this differs from controls.
In work package 1, I will also study why other sea urchin species were unaffected during the 1983 and 2022 die-off event. Researching why these species were not infected and how their immune systems differ from D. antillarum, could be crucial to discover why D. antillarum is more susceptible to these die-off events. I will repeat the exposure experiments with adults of two other sea urchin species, namely: a) a sea urchin species that co-existed with D. antillarum in the Caribbean and was unaffected during the 1983 and 2022 die-off events, and b) another Diadematidae species that did not occur in the die-off area to study whether susceptibility is species- or family specific.
Work package 2: Population dynamics and recovery of D. antillarum
The massive die-off events in 1983 and 2022 greatly reduced the D. antillarum population size which may have reduced the genetic diversity unless its long pelagic larval phase has supported gene flow through population connectivity. Larval dispersal through the Caribbean and between populations is heavily understudied, meaning we do not know where larvae end up and where they come from (also called sink- and source populations). Studying the larval dispersal and population connectivity enables assessment of prospects for future (assisted) recovery of D. antillarum. In addition, locations identified as important for larval dispersal and connectivity could be protected to safeguard higher overall natural recovery success and to target for restocking by restoration efforts.
To study the population connectivity and larval dispersal genetic diversity will be determined in tissue samples and larvae of wild D. antillarum populations collected from various locations in the Caribbean. The results of this research will be input for the modelling work of PhD candidate Pam van Touw as well as provide validation for the modelling results. In this model, genetic diversity will be combined with particle movement modelling to simulate larval dispersal and identify how populations are connected within the Caribbean. Additionally, this model will be used to simulate how larval dispersal and population connectivity would be affected under future scenarios such as climate change, anthropogenic activities, and new pathogen outbreaks. This way, we will gain insights in prospects for resilience and future recovery of D. antillarum populations.