
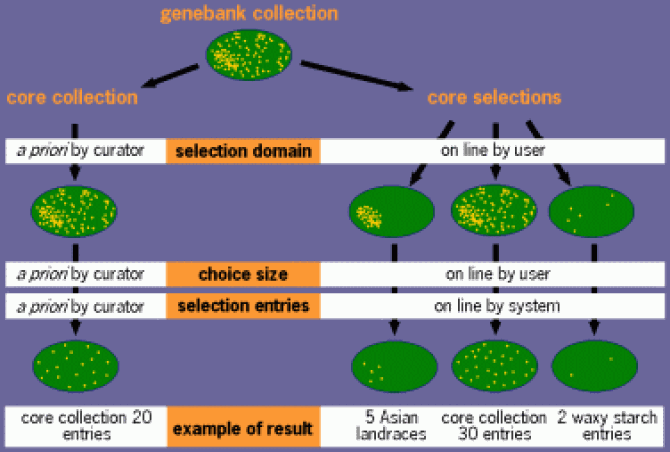
Core selections
Where core collections are fixed sets of accessions, core selections are created answering a specific request of a user who specifies the size and domain of the selection.
The general procedure for the selection of a core collection generally includes a number of steps
- The material that should be represented, i.e. the domain of the core collection, has to be defined.
- The domain has to be divided in distinct groups.
- The number of entries per group has to be decided.
- Entries from each group have to be chosen.
In a "classical" core collection these steps are taken a priori, the product being the core collection. For a core selection, only the first two steps are fixed. Although some default values are available, the decision about how many accessions from each group, and which accessions to include, can be taken by the user.
The idea
Domain
The first step, the definition of the domain, is fairly straightforward. Basically, it can be any set of germplasm accessions. As an example, the CGN lettuce collection will be used, but also butterhead lettuce with yellow leaves could have been used.
Path indicator
The division into distinct groups is achieved using a stepwise procedure (stratification) resulting in an hierarchy, which can be graphically represented by a "diversity tree" . Firstly, the important major divisions are made, followed by splitting these subgroups into smaller ones, etc. This splitting into subgroups is continued until the subgroups are genetically homogeneous or there is no information to base further groupings. Groups that cannot be divided any further are called end groups.
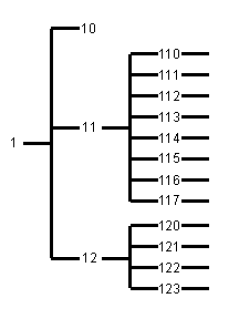
The hierarchy is recorded in a descriptor called the "path indicator". This path indicator is a series of ciphers which describe the subsequent divisions. This can best be explained using an example. In the case of the CGN lettuce collection, all path indicators started with a 1 indicating that it was part of the lettuce collection. A first distinction was made between the groups: 11 - cultivated Lactuca, 12 - non-cultivated material and a residual group 10 - unknown whether cultivated or not. Within group 11, eight subgroups were defined: 111 - Butterhead lettuce, 112 - Cos lettuce, 113 - Crisp lettuce, 114 - Cutting lettuce, 115 - Latin lettuce, 116 - Stalk lettuce, 117 - Oilseed lettuce, and 110 - unknown cultivation type. Within group 12, four subgroups were defined: 121 - primary genepool (L. serriola), 122 - secondary genepool, 123 - tertiary genepool, and 120 - genepool unknown. Within most of these groups further divisions were defined. For example, all accessions of L. quercina ended up in group 123131, which was a subgroup of group 12313 - section Lactucopsis, subgroup of 1231 - Lactuca species in 123 - the tertiary genepool of 12 - non-cultivated material from 1 - the total collection.
Allocation
As a starting point for the division of entries into groups, a default weight has to be defined for all subgroups within each group. This weight should correspond to the relative importance of the subgroups. There are several ways of deciding on this relative importance. If there is no further knowledge about the subgroups, one can decide about the importance on the basis of the number of accessions in the group. If sufficient material in the groups has been characterized with genetic markers it is possible to compare the marker diversity within the subgroups and base the weight on this comparison. Finally, if the importance of the group to the user should be a factor, or if there is a general idea about the genetic diversity in the groups, a subjective decision can be made about how many entries should be allotted to each subgroup. This final option is usually the easiest and most effective.
Priorities
To assist in the choice of entries per endgroup, a priority for inclusion in the core selection has to be given to each accession; accessions with the highest priority will be included first.
As a starting point, these priorities can be given randomly. But it is preferable to determine the priority on the basis of further available information. For example, it is possible to create an index on the basis of availability of passport or characterization data; an accession without country of origin, or a variety without a name should be given low priority; accessions with much characterization data should be given high priority. It is also possible to give higher priority to accessions with a high reputation that played an important role in breeding history, for example, or that are being used as a standard in research. Or, finally, it is possible to assign the priorities in such a way that the accessions chosen will represent the diversity in the endgroup best. For example, if data on flower color are available and three colors exist, the three highest priorities might be assigned to accessions with different colors. If many data are available, one might consider a multivariate approach to this prioritizing.
The method
A core selector has the following basic elements:
- Data structures storing (1) the hierarchy and priorities of accessions within the collection, and (2) the relative default importance of the different groups in the collection.
- An interface which allows the user to indicate the desired size of the core selection, and to change the default distribution of entries in a group over its subgroups.
- Algorithms which make the final selection, and produce the output.
Data structures
The data structures can be created in any environment, though tables in the database containing the rest of the information about the database will generally be preferable.
- Table PATHS contains information about the groups and their relative importance. It consists of three columns:
- PATH (the unique key) which contains the path indicator as described above, GROUP which contains a short description of the group, and WEIGHT which contains the default relative importance of the groups (see Table 1).
The weights used in this example are directly derived from a previously defined core collection from the CGN lettuce collection.
- The second table, ANR_PATH, contains information about the relation between accessions and groups. It consists of three columns: ANR (the unique key) the accession number, PATH which contains the number of the group the accession belongs to, and PRIORITY which indicates the order in which accessions within a group should be included in the core selection. (Table 2) Columns PATH and PRIORITY might also be added to another table with the accession number as unique key, such as the table containing passport data.
User interface
When a user enters the system, information about the starting point of the core selection should be displayed, such as the number of available accessions. The user is then prompted to enter the approximate size of the core selection. Next, the user is presented with the first division in subgroups, the number of accessions and the default number of core entries in each subgroup. At this point she/he has three options: 1 - accept the allocations, 2 - alter the default number of entries in the various subgroups, 3 - select one or more subgroups which she/he would like to define further. If the third option is chosen, the subgroups within the selected group, plus numbers of accessions and the default numbers of core entries in each subgroup are presented. This option can be followed for all subgroups defined within the selected group. Once the sizes of all groups have been determined up to the level desired by the user, the core selection can be generated.
The resulting request, at this level could look like: '50 accessions representing the West European cultivars of butterhead lettuce, 50 accessions representing all other butterhead lettuce in the world, and finally 25 accessions representing the other types of cultivated Lactuca'. The ideal situation would be to create an extra layer over the user interface of a genebanks documentation system allowing the user, possibly via the Internet, to:
- Define and refine the requirements of the material to be requested, and if the number of accessions meeting the requirements would be to large to:
- Adjust the default distribution over the types of material within the selection in a user friendly manner.
- Inspect the selected accessions, and possibly adapt and fine-tune the selection.
- Order the selected material from the genebank.
Algorithms
To divide the entries in a group over the subgroups within this group, the system uses the following principles:
- There cannot be more entries in a group than there are accessions of the respective group in the collection.
- Entries in a group are divided over the subgroups using the values of WEIGHT as weighting factors. Entries in a subgroup with a WEIGHT equaling zero will only be selected if all accessions in the other subgroups have been sampled completely.
- Apart from the groups with a WEIGHT equaling zero, all groups should be represented with at least one entry. If this is not possible since there are more subgroups than entries, the entries are allotted to the subgroups with highest values for WEIGHT.
Examples of the allocation of entries over groups given the number of accessions in the group and the relative weights of the groups are presented in Table 3.