
Project
Genome-wide metabolic modelling and data integration of organic acid production in filamentous fungi
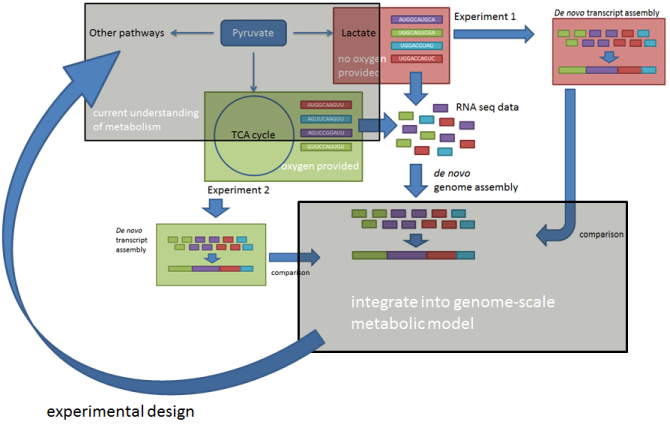
Despite the wide use of filamentous fungi for industrial organic acid production, the underlying metabolic processes are not yet well known and understood. Semi-random overexpression or knockouts of genes known or expected to be involved in the pathway can lead to more efficient and hence cost-effective production of the desired organic acid, however a better understanding of the pathway can lead to more directed approaches in metabolic engineering.
Data driven modeling can lead to the desired level of understanding, which in turn can lead to better experimental design and execution and thus save time usually spent for the trial and error cycle. In the given project, data from organic acid production in filamentous fungi is collected and integrated into genome-scale metabolic models. Compartmentalization and the intracellular pH of the organism studied will be taken into consideration in order to lead to a model able to allow targeted modifications in the pathway of the organic acid of interest. Additionally, pathways which are not endogenous in a given filamentous fungus can be implemented by integrating the necessary genes from another fungus into the genome. The focus will lie on the production of citric, itaconic and/or fumaric acid in A. niger, which is a widely known and accepted industrial working horse for the production of citric acid.
WCSB
This project is part of the strategic plan of Wageningen UR about Systems Biology for Food, Feed, and Health. More information about WCSB and the other 13 projects involved can be found at the website www.wur.nl/systemsbiology.