Thesis subject
Characterization of an ultra-conserved angiosperm sequence motif
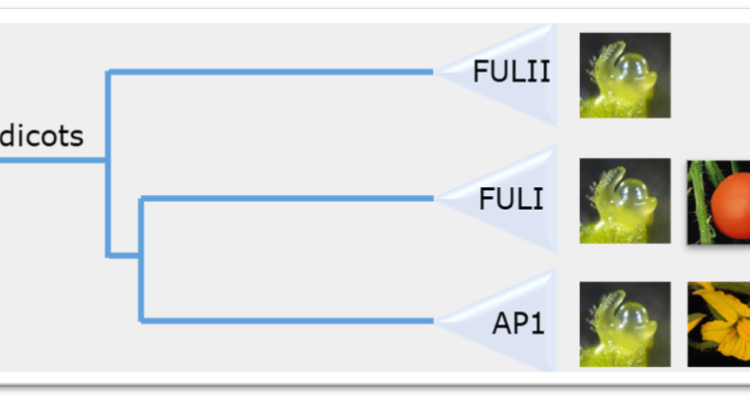
Transcription factors that belong to the FRUITFULL/APETALA 1 clade are essential regulators of flowering time, flower initiation and fruit development. In the eudicots, the FUL/AP1 genes have diverged and both gained functions (neo-functionalized) and divided functions (subfunctionalized). This is apparent from their expression patterns and linked to divergence in the regulatory region of the genes. We identified an ultra-conserved regulatory motif, named R1, which plays a role in meristem expression and fruit expression, and is linked to the functional divergence. In this project, we aim to find out how the sequence divergence in this motif can be linked to the observed expression divergence. In this stage of the project, we predominantly aim to identify the upstream transcription factors that can bind to the R1 motif.
This involves:
- Testing expression in different tomato lines: RNA extraction, cDNA synthesis, qPCR
- Test binding of transcription factors to DNA using EMSA.
- Transient luciferase assays to test which transcription factors activate or repress R1.
- Hormone treatments to test the responsiveness of R1 to hormones
- GUS reporter assays
Requirements:
Interest in the project and basic knowledge of molecular biology.