Project
Phylogenomic synteny analysis of plants and animals
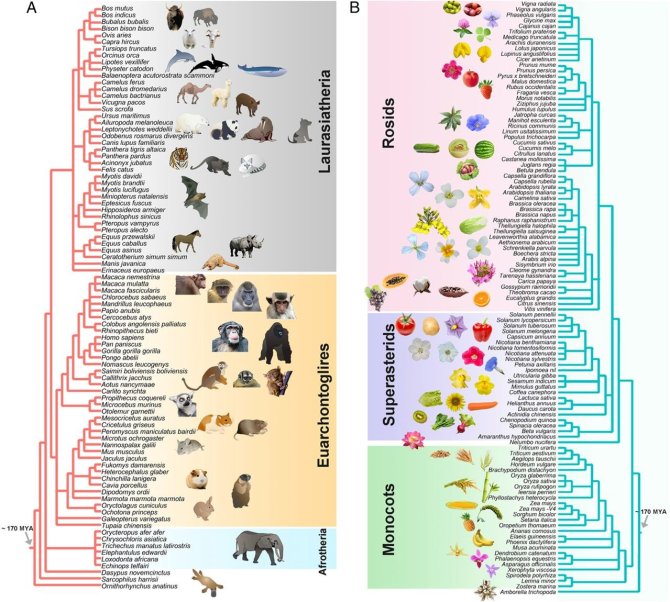
Studying the organization of genes within genomes across broad evolutionary timescales can advance our understanding of the evolution of traits and clades. We use network approaches to investigate genome dynamics of angiosperms, mammals and birds. In general, genome organization and gene microcollinearity is much more conserved in birds and mammals than in flowering plants. We then identified the genomic outliers or “rebel genes,” within each clade.
Genes that have moved are unusual for mammals, whereas highly conserved single-copy genes are exceptional for plants. How conservation and changes in synteny or fundamental differences in genome organisation have contributed to the evolution of lineages could be a new scientific frontier.
Publications
-
Data for manuscript "Synteny identifies reliable orthologs for phylogenomics and comparative genomics of the Brassicaceae"
-
syntenet: an R/Bioconductor package for the inference and analysis of synteny networks
Bioinformatics (2023), Volume: 39, Issue: 1 - ISSN 1367-4803 -
GENESPACE tracks regions of interest and gene copy number variation across multiple genomes
eLife (2022), Volume: 11 - ISSN 2050-084X -
Whole-genome microsynteny-based phylogeny of angiosperms
Nature Communications (2021), Volume: 12, Issue: 1 - ISSN 2041-1723 -
Network-based microsynteny analysis identifies major differences and genomic outliers in mammalian and angiosperm genomes
Proceedings of the National Academy of Sciences of the United States of America (2019), Volume: 116, Issue: 6 - ISSN 0027-8424 - p. 2165-2174.