Project
Internship:Democratisation and FAIRification of standardised Anti-Microbial Resistance (AMR) detection workflows in a Virtual Benchmarking platform (VBP).
Group: Pathology and Clinical Bioinformatics, Erasmus MC, Rotterdam (EMC)
Background and research interests - The current endemic threat of antimicrobial resistance (AMR) is a huge global problem, impacting on individual and societal health, national economics and healthcare system sustainability. Tackling AMR globally requires global action: reducing inaccurate antimicrobial use, implementing infection prevention strategies, preventing antimicrobial contamination of foodstuffs and the environment, development of alternative antimicrobial therapies, and rapid diagnostics to detect microbial pathogens and their AMR characteristics. In that respect, next generation sequencing (NGS) methods are being widely used to sequence the genomes of microbes in order to obtain information on the genetic relatedness of outbreaks of infection and also to predict AMR phenotypes from genotypic information.
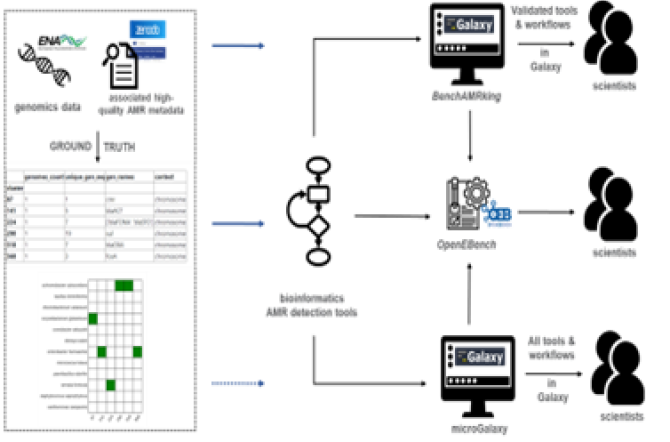
However, to date there is a lack of a consolidated virtual benchmarking platform (VBP) that could provide the first step in facilitating such activities - therebytranslating theory into practice. The B2B2B AMR DX JPIAMR network,
https://www.jpiamr.eu/projects/b2b2b-amrdx/ is a cross-disciplinary, geographically diverse and gender-balanced group of partners from universities (bench), hospitals (bedside), for-profits (business), governmental and nonprofit organisations (beyond), to develop an Antimicrobial Resistance-Virtual Benchmarking Platform (AMR-VBP). EMC (Dr Stubbs, Dr Hays & Mr. Dollee) have developed a BenchAMRking (https://erasmusmc-bioinformatics.github.io/benchAMRking/ ), a prototype AMR-VBP.
Objectives
The aims of the study are to:
- Complete the development of BenchAMRking application
- Extend the application with long-read and/or plasmid sequence benchmarking activities
Methodology / what students can learn
The student will extend the existing VBP BenchAMRmarking platform.
The student will learn to and be expected to develop, execute, manage, report and complete the research plan developed in collaboration with EMC.
Requirements
We are looking for Bioinformatics MSc students interested in developing their analytical research skills more advanced analytical techniques. The student should be proficient in python and R. The project will have a duration of 6 months.

Contact information
Andrew Stubbs, Pathology and Clinical Bioinformatics – a.stubbs@erasmusmc.nl
John Hays, Medical Microbiology - j.hays@erasmusmc.nl
Peter van Baarlen, Host-Microbe Interactomics, Wageningen University Animal Sciences - peter.vanbaarlen@wur.nl