Project
Building a robust PVC species phylogeny
This project aims to decipher the evolutionary histories of PVC bacteria and investigate the evolution of their genome content underlying their unique cellular biology.
Background
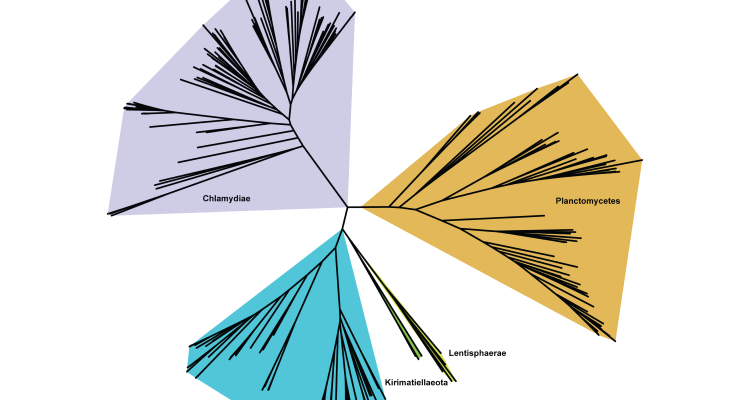
The PVC (Planctomycetes–Verrucomicrobia–Chlamydiae) superphylum is a group of bacterial phyla, the members of which often exhibit unique cell biological attributes and a well-supported monophyly. In recent years, however, the PVC superphylum has experienced a major expansion concerning the microbial diversity it comprises mainly due to the advent of next-generation sequencing and metagenomics. This has led to the accumulation of novel microbial groups that, depending on the phylogenomic datasets and methods used, either form a monophyletic relationship with traditional PVC members or appear as sister lineages. Since this has resulted in phylogenetic inconsistencies, there have been conflicting descriptions of the phyla comprising PVC. As such identifying the true members of PVC bacteria and their evolutionary relationships is of great importance.
Aim of the project
The aim of this study is to develop and employ large-scale phylogenomics that will account for sources of error from different key steps of phylogenetic analyses as well as strategies to mitigate these. This will not only contribute to inferring more accurate phylogenetic relationships among PVC members, but also to building a comprehensive evolutionary framework where we can reliably investigate the evolution of their genome content underlying their unique cellular biology.
BSc/MSc theses
This project is available for BSc or MSc students interested in phylogenomics and comparative genomics. Experience with working on linux command line and programming (e.g., python, R) is appreciated. Please contact me via the contact form.